Breadcrumb
- Home
- Research
Research
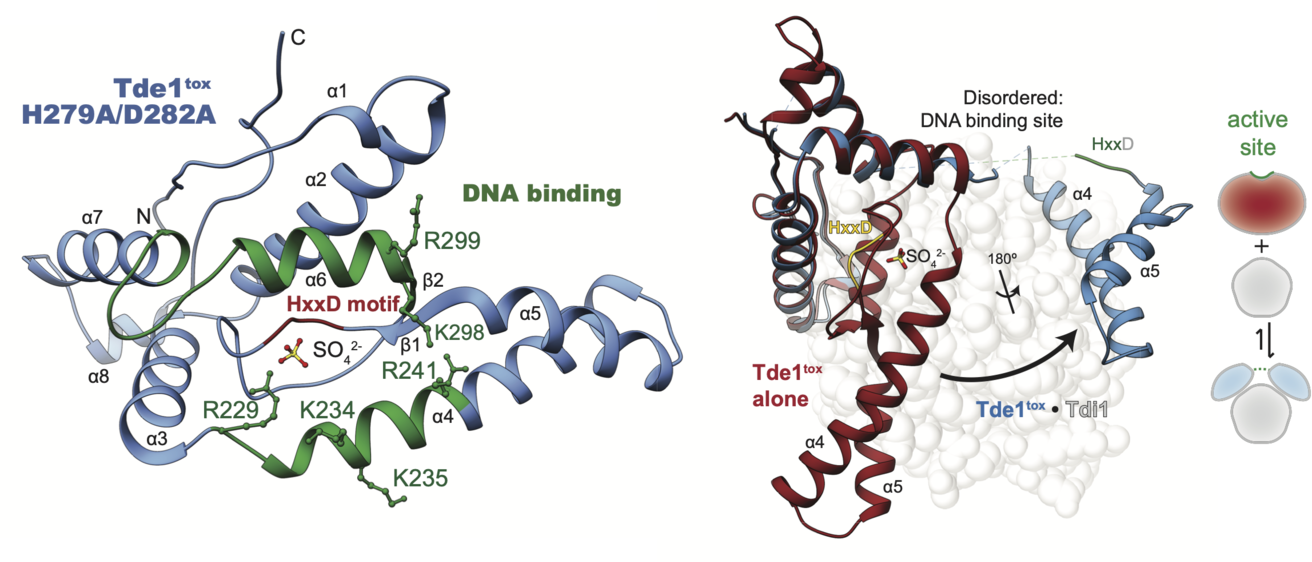
Structural disruption of Tde nuclease effector domains by immunity proteins
Bacteroidales use type VI secretion system DNAse effectors (Tde) for interbacterial competition. A crystal structure of catalytically inactive Tde highlights a DNA binding site (green) and enzymatic active site with conserved HxxD motif. Immunity protein (Tdi) interaction induces a large effector conformational change in Tde nucleases, disrupting the active site and altering the DNA binding site, highlighted by a Tde / Tdi complex crystal structure. The Tde/Tdi interface and immunity mechanism are distinct from all other polymorphic toxin–immunity interactions of known structure. Tde encoding genes are enriched in fecal metagenomes from ulcerative colitis patients, suggesting a link to inflammatory bowel disease.
Reference:
Bosch DE, Abbasian R, Parajuil B, Peterson SB, Mougous JD. Structural disruption of Ntox15 nuclease effector domains by immunity proteins protects against type VI secretion system intoxication in Bacteroidales. mBio. 2023. Epub head of print. doi: 10.1128/mbio.01039-23
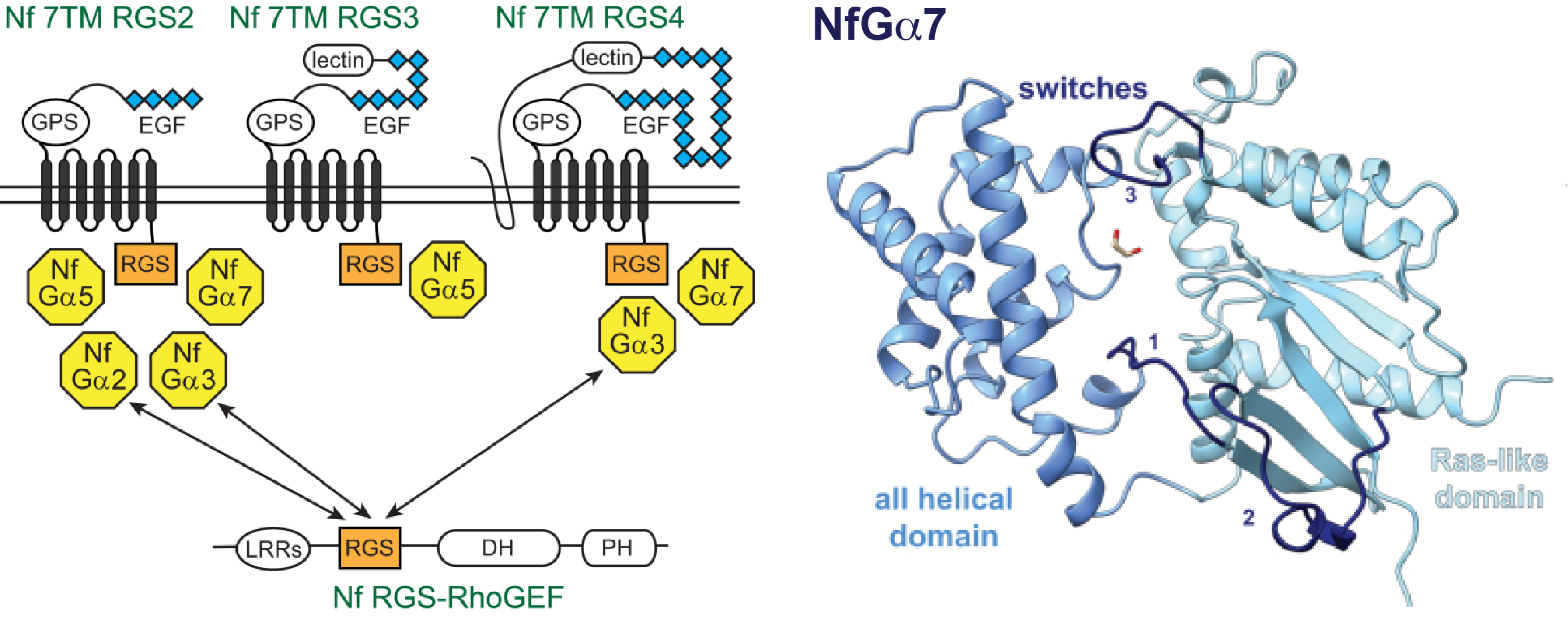
Heterotrimeric G protein signaling in Nagleria fowleri
The free-living amoeba Naegleria fowleri is an uncommon human pathogen with infection mortality rates near 100%. Primary amoebic meningoencephalitis arises when N. fowleri from contaminated water sources enters the nasal cavity and ascends along olfactory nerves to the brain. Our laboratory identified heterotrimeric G-protein signaling pathways that may mediate response to extracellular cues that may be targetable with small molecule drugs. The single celled N. fowleri expresses an array of GTPase G-alpha subunits, some of which are "self-activating" through rapid nucleotide exchange mechanisms also observed in plants and some protozoa. A crystallographic snapshot structure of the self-activating NfG-alpha7 captures a nucleotide-free state. G-alpha subunits engage regulators of G-protein signaling (RGS proteins), including an RGS domain-containing Rho GTPase guanine nucleotide exchange factor (RhoGEF). We hypothesize that these signaling pathways mediate cytoskeletal rearrangements in response to extracellular stimuli, for example during amoebic motility.
Reference
Bosch DE, Jeck WR, Siderovski DP. Naegleria fowleri conventional and self-activating G protein alpha subunits engage seven-transmembrane RGS proteins and an RGS-RhoGEF effector. J. Biol Chem. 2022 June. 20:102167. PMID 3578399.

Tumor microbiomes in IBD colitis-associated carcinoma
Dysbiosis and pathobionts contribute to inflammation and the risk of colitis-associated carcinoma (CAC) in animal models, but their roles in humans with this uncommon disease are unknown. We identified microbiome differences in human CAC compared with longstanding inflammatory bowel disease (IBD) and sporadic colorectal carcinoma (CRC). Twenty-four CAC resections were matched with CRC and IBD controls. Methods included histopathology, 16S rDNA metagenomics, and pathobiont-specific qPCR. Beta diversity differed by diagnosis (PERMANOVA p = 0.007). The distinguishing taxa included Akkermansia enriched in CRC, and Bacteroides spp. enriched in IBD. The non-neoplastic mucosae presented distinct beta diversity (p = 0.005), but the CAC/CRC tumor microbiomes were similar (p = 0.7). Within metastases and margins, Enterobacteriaceae were enriched in CAC, and Bacteroidales in CRC. Pathobiont-specific qPCR confirmed a greater frequency of pks+ E. coli and enterotoxigenic Bacteroides fragilis in CAC than IBD. High alpha diversity was associated with active inflammation, advanced cancer stage, and shorter overall survival (log-rank p = 0.008). Mucosal microbiomes distinguish CAC from longstanding IBD, implicating pathobionts as markers for disease progression. Integrating our findings with prior animal model research, pathobionts promote carcinogenesis in IBD patients through genotoxicity and host cell signaling.
Reference
Masaadeh AH, Eletrebi M, Parajuli B, De Jager N, Bosch DE. Human colitis-associated colorectal carcinoma progression is accompanied by dysbiosis with enriched pathobionts. Gut Microbes. 2025 Dec;17(1):2479774. doi: 10.1080/19490976.2025.2479774. Epub 2025 Mar 17. PMID: 40094201; PMCID: PMC11917176.
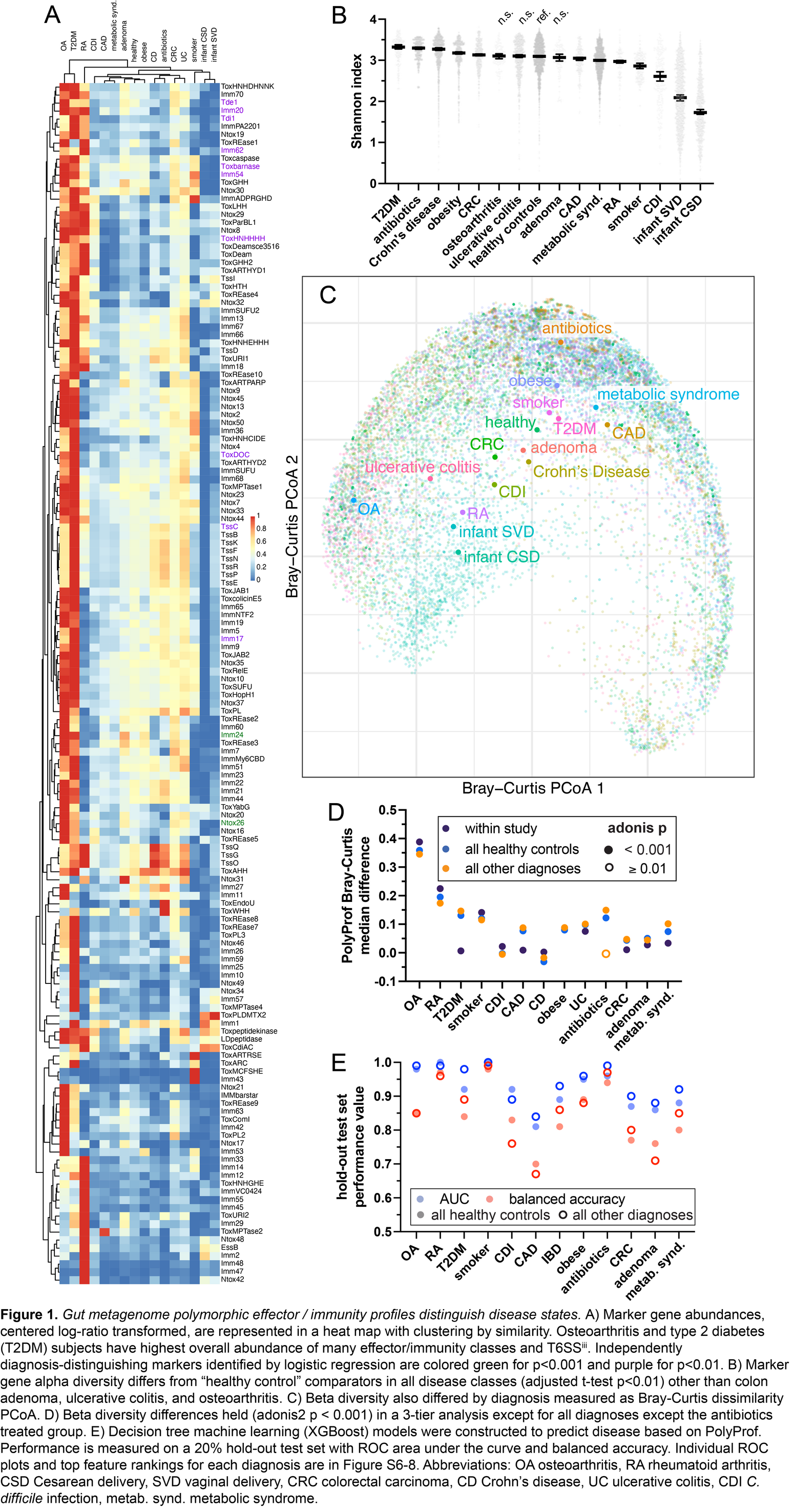
Metagenomic polymorphic toxin effector and immunity profiling
Bacteria use antagonistic interbacterial weapons such as polymorphic toxin secretion systems (TSS) to compete for niches in the human gut microbiome. We developed a bioinformatic marker gene approach (PolyProf) to quantify TSS including ∼200 effector and immunity genes and applied it to ∼15,000 publicly available human metagenomes. PolyProf alpha and beta diversity readily distinguished 12 different human disease states. Decision tree machine learning models integrating bacterial taxonomy with PolyProf had near-perfect accuracy (ROC area 1.00) for all 12 disease states. During microbiome development in the first year of life, PolyProf alpha diversity increases, and beta diversity becomes increasingly like the maternal microbiome, influenced by vertical transfer, delivery mode, and breastfeeding. PolyProf is related to strain sharing among adults through social interactions. In summary, interbacterial antagonism with TSS shapes microbiome development and interpersonal strain sharing. Since PolyProf distinguishes diverse adult disease statuses, these dynamics may contribute to non-genetic inheritance.
Reference
Schroer HW, Beghini F, Garay JAR, Christakis NA, Bosch DE. Metagenomic polymorphic toxin effector and immunity profiling predicts microbiome development and disease-related dysbiosis. bioRxiv [Preprint]. 2025 Jul 8:2025.07.08.662037. doi: 10.1101/2025.07.08.662037. PMID: 40672213; PMCID: PMC12265584.